The Biophysical World Inside a Jam-Packed Cell
Introduction
It’s a familiar image, reprinted in countless biology textbooks: an illustration of a typical cell, halved like a grapefruit to reveal its innards. Strands of endoplasmic reticulum encircle a nucleus that floats in the center like a raft. RNA molecules wait patiently at ribosomes to deliver recipes for making proteins. A few vacuoles and Golgi bodies bob about. A mostly deserted cytosol offers a blank backdrop. These are scenes of a calm, rarefied order, as if a cell were a tidy factory with workers individually going about their focused tasks.
Scrub that picture from your mind right now: Cells are anything but calm or rarefied. If you were trapped inside a cell, you would feel like a dancer in a thronged nightclub, constantly jostled by neighbors.
Over the past few years, thanks to stunning advances in imaging and genetic engineering, scientists have been able to observe and measure crowding inside cells in living organisms for the first time. The experiments have revealed a more dynamic and crowded place than anyone expected, and are the latest evidence that cells actively regulate their internal crowdedness to optimize for the chemical reactions required for life.
Far from being a biophysical accident, the packing of molecules into tiny spaces is emerging as a fundamental way that cells have evolved to harness physics to bring molecules together for the estimated 1 billion biochemical reactions that occur every second in every cell of our bodies.

The cell biologist Liam Holt pioneered a method that combines genetic engineering and fluorescence microscopy to trace ribosome-size molecules moving through a cell.
Deb Bemis
“If you’re in a bar or nightclub that’s not very crowded, you might not talk to anyone or dance with anyone,” said Liam Holt, a cell biologist at New York University Langone Health. “But at midnight when the bar is jam-packed, you’re more likely to talk or dance with someone next to you. But if you see someone across the room, it’s harder to get to them.”
The new findings are confounding scientists’ expectations, raising questions about how exactly molecules can encounter their reactive partners in a teeming, crowded space — and therefore how cells can possibly function.
The Biophysical Knife’s Edge
Cells may be biological entities, but they are not exempt from the laws of physics. In his 1944 book What Is Life? The Physical Aspect of the Living Cell, the quantum physicist Erwin Schrödinger argued that living things, like their nonliving counterparts, must submit to governance by physical laws. His vision has inspired biology-minded physicists and physics-minded biologists ever since.
But studying the physics of eukaryotic cells — the kind that make up our bodies and those of other multicellular organisms — presented a challenge: How do you study an individual cell buried deep within the body of a person, a mouse, or even a simple worm?
At first, scientists got around this problem by removing those cells from humans and animals and growing them in test tubes or petri dishes. Early studies hinted that cells are subject to a Goldilocks phenomenon: They function best when their cytoplasm — everything enclosed by a cell’s membrane, including organelles, molecular structures such as ribosomes, and the gel-like cytosol and its dissolved molecules — has some level of crowding, but not too much. In the 1980s, a team of researchers found that if they diluted the cytoplasm extracted from frog eggs even a little bit, vital biochemical reactions such as mitosis and DNA replication ceased. Other studies found that overcrowding could be equally disastrous, causing the chemical machinery of life to freeze up.
Cells are constantly churning through energy to stir things up, keep the cytosol fluid, and encourage molecules to collide and react more often than they would through simple diffusion. Even so, cellular life appears to balance on a knife’s edge. If cells were any less crowded, molecules would wander aimlessly and only rarely encounter their partner (or partners) in the chemical reactions that power life — metabolism, protein synthesis, growth, division, and more. In that situation, cellular life would wither. If, on the other hand, cells were much more crowded, molecules would be stuck in place, unable to move much at all, let alone come across their reaction partners. Life would grind to a halt.
Evolution seemed to have struck a delicate balance between over- and undercrowding, with large molecules such as ribosomes typically representing between 30% and 40% of the volume of dissolved macromolecules in the cytosol, Holt said. “It seems that much of biology is tuned to have a very similar level of crowding.” But to confirm this view, researchers would need to find a way to track molecules moving through a cell. They would need a tracer of the right size.
Crowd Control
Crowding is relative. While a person might have trouble moving through Holt’s imagined club at midnight, a cat or mouse would not find it too crowded to navigate. To study crowding in cells, biologists needed a proxy molecule in the same size range — a tracer roughly as large as the large molecules involved in most cellular reactions.
In the mid-2010s, Holt introduced genetically encoded multimeric nanoparticles, or GEMs, which are naturally occurring spherical proteins about 40 nanometers in diameter — around the same size as ribosomes, the molecular machines that build proteins. Using genetic engineering, researchers can decorate the surfaces of GEMs with glowing green fluorescent tags and then track their movements through a cell’s cytoplasm under a microscope.
In 2018, this approach gave Holt and his colleagues fresh insight into how cells manage their crowdedness. They put GEMs inside yeast and human cells in culture and measured how long it took the particles to percolate through different areas of the cell. Strangely, in cells grown under different nutritional conditions, the crowding of the entire cell seemed to change. “This led me to ask what was going on,” Holt said.
He suspected that mTORC1 was involved. The main nutrient sensor in eukaryotic cells, mTORC1 is a master regulator of cell growth; based on nutrient levels, it can boost production of ribosomes to build more proteins faster. “The rate at which organisms can grow is fundamentally limited by how many ribosomes they can produce,” Holt said. Indeed, when his team chemically suppressed mTORC1, the concentration of ribosomes decreased, and GEMs flowed through the cytoplasm much more easily.
Further experiments suggested that cells can use ribosomes — one of the most abundant molecules in a cell — and the genetic pathways that regulate them, to control their molecular crowds. Holt calls mTORC1 “a dynamic control knob for cytoplasmic physical properties.”
Holt’s team’s work on how cells manage their internal environment inspired further experiments. The paper, now cited almost 500 times, was “incredibly influential,” said Arohan Subramanya, a cell biologist at the University of Pittsburgh. Holt showed that “ribosomes act as natural crowding agents.”
The result is a biochemical system in which the crowdedness of the cytoplasm reflects a cell’s growth and health, Holt said. “If organisms are growing quickly and in perfect conditions, they pack their cytoplasm with a very high concentration of ribosomes.”
However, his team made their discovery in single-celled yeast and cultured human cells. G.W. Gant Luxton, a biophysicist, wanted to see if cells followed the same rules in the more complex environment of a living multicellular organism.
Jam-Packed
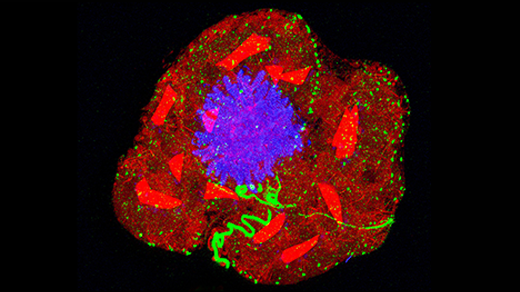
In summer 2018, Luxton and Holt met for coffee while both were visiting the Marine Biological Laboratory in Woods Hole, Massachusetts. It took only a few minutes for Luxton to see how to use Holt’s GEMs in his work — but he would need more help. He contacted a colleague, the geneticist Daniel Starr, to see if he would be interested in engineering the particles into Caenorhabditis elegans, a microscopic worm. Crucially, this workhorse model organism is transparent, allowing biologists to observe experimental fluorescence such as that given off by GEMs.
Starr said yes. After receiving funding for the project, Luxton moved his lab to the University of California, Davis, to form the Starr-Luxton lab. It took months for Luxton and Starr to make the worms produce GEMs, and years to figure out how to image and analyze the data. But eventually they could see and measure the particles glowing in the worms’ gut and skin cells.
Once inside the cells, the particles hardly moved. The worm cytoplasm, measurements showed, was around 50 times more crowded with ribosomes than that of Holt’s cultured cells. At first, the graduate student conducting the experiments thought they had made a mistake.
“It just kind of blew my mind,” Luxton said. “Why aren’t these probes moving?”
Another graduate student remarked that while cultured cells had the consistency of honey, the thicker worm cytoplasm resembled “strawberry jam.”
It was “a very surprising result,” said Holt, a co-author on the paper, which was published in September 2025 in Science Advances. “I’ve never seen anything as dramatic as in the epithelium of the worm.”
For Luxton, the result immediately raised questions. For one thing, if the interior of living cells is thick like jam, how does a given molecule move around enough to encounter another molecule it needs to react with? “I don’t know how anything ever finds anything,” he said.

Xiangyi Ding (left), G.W. Gant Luxton (center), and Daniel Starr (right) engineered GEMs into transparent worms. The cytoplasm of the multicellular animal was 50 times more crowded than that of cells grown in culture.
Joaquin Benitez, UC Davis
The researchers also noticed that the GEMs seemed stuck in certain regions of the cell. But when Luxton and Starr’s team disrupted the functioning of a large protein called ANC-1 that acts as scaffolding inside cells, the GEMs started moving. This suggested that cells have evolved multiple mechanisms to manage crowding in the cytoplasm.
“Think of the cell as a box and ribosomes as packing peanuts,” Luxton said. “You can change crowding through the packing peanuts … but you can also change how big the box is. It looks like ANC-1’s controlling that aspect of this.
“It shows there’s very different ways to control crowding,” he continued. “Cells in tissues may depend much more on spatial organization — compartmentalization, scaffolding, channeling of substrates between enzymes — than we appreciate from studying cells in culture.”
A New Subfield
Since the publication of the paper on ANC-1, Luxton and his team have put GEMs in worm neurons and other kinds of cells, including diseased and aging ones, to gather baseline data on the cytoplasmic biophysical properties of different tissues. “We’ve been building an atlas of worms,” he said. With a collaborator, they’ve also started to put GEMs in zebra fish, another common model organism. They are finding a range of crowdedness levels within cells, complicating Holt’s picture that cells prefer to fill 30% to 40% of their volume with molecules. The research underscores the importance of confirming cell culture results in living organisms, which operate under different conditions.
“Cells have found different ways to deal with a continuum of crowdedness,” Luxton said. “It really matters where you’re looking, and not all tissues are created equal.”
Holt agrees. “Rather than one universal optimal crowding level, it appears that different cell types and tissues tune their crowding to suit their particular needs,” he said. It makes sense that, say, a muscle cell that needs to repeatedly contract and relax would have different mechanical properties than a fat cell whose main job is to store energy.
Another line of research involves putting GEMs in organoids — three-dimensional lab-grown structures that can be made to mimic various tissues and organs. Because organoids are 3D, Luxton believes they better approximate living animals than cells floating in test tubes. He and colleagues are putting GEMs into pancreatic cancer organoids and looking for biophysical differences that could be used to distinguish cancer cells from healthy ones.
Scientists have long known that cancer cells are physically distinct. “Cancer is an example where you have big mechanical changes,” Holt said. “The way you find a tumor, you look for a lump — a larger cell mass than should be there. It’s like pumping air into a tire. Cells are getting squished; they get more crowded.” This, he suggested, would change their biophysical properties.
It’s a heady time, with innovations in biophysics, microscopy, and genetic engineering coming together to open up a new subfield within the centuries-old science of cells.
“It’s kind of like Pandora’s box,” Luxton said. “Every time we look at a different tissue, we see something we don’t expect.”